Bacteria Identification from Given Sample Skills: Organism Isolation and Antimicrobial Sensitivity Testing

Question for bacterial identification
For bacteria identification of organisms, you can ask if you have any queries related to a given question.
A broth culture is given to you to isolate and identify the organism and also produce its antibiotic sensitivity profile by Saturday. Please do not proceed to another experiment before showing the result of your experiment to your teacher.
Time of inoculation – 1: 00 pm
Type of broth – BHI
Incubation – aerobically
Temperature –?
Isolation from where?
Mixed /single -?
Experimentation Day 1 (Year -month -day i.e. Thursday) for bacterial identification
I was given a broth Labelled as ‘ k’ at 1:00 pm in a broth.
I incubate the broth at 37 ºC for 4 hours aerobically to obtain the growth of the given organ.
Both media BHI (Brain Heart Infusion ) broth.
No added factors/ingredients
After 4 hours (5:00 pm)
Broth shows
- Uniform turbidity
- No granular formation
- No pellicle formation
- No pigment formation seen
- No gas formation
Gram stain for bacteria identification
Performed on broth with
Positive Control – Staphylococcus accreus ATCC25923
Negative control – Escherichia Coli ATCC 25922
Result of Gram’s stain
Negative control shows – Gram-negative bacilli
Positive control shows -Gram-positive Cocci
Test organism: Gram-negative bacilli
size 1-3 µm x 0.4 – 0.7 µm
Rod-shaped, single, straight rounded ends with parallel side
Arranged singly
No pleomorphic forms seen
No evidence of spore
No evidence of capsule
No mixture pure form seen
Hanging drop preparation for bacteria identification
It shows actively motile organisms.
Possible Pathogens organisms
Escherichia
Salmonella
Proteus
Providencia
Morganella
Pseudomonas
Enterobacter
Burkholderia
Stenotrophomonas
Vibrio
Aeromonas
Plesiomonas
Serratia
Moraxella
Haemophilus
Hafnia
Further Processing
Further, it was processed for isolation and indention; inoculation of broth on
MacConkey agar
Blood Agar
Chocolate
Inoculation By streaking method and plates were incubated at 37 °C aerobically overnight at 5 pm.
Day 2 for bacterial identification
Colony characteristics were studied at 11:00 am from all three plates in transmitted bright light which is helpful for bacteria identification to some extent.
Gram Staining from colonies of three plates
Nutrient Agar : Gram-negative bacilli , 1-3 x 0.4 -0.7 µm
MacConkey agar – Gram-Negative bacilli
Blood Agar – Gram-Negative Bacilli
All three media show similar types of colonies/bacteria in Gram’s stain
Biochemical Tests for bacteria identification
Catalase test for bacteria identification
Positive control – Staphylococcus auress ATCC 25923
Negative control –Enterococcus faecalis ATCC 29912
Test organism – Shows bubbling when adding 3 % H2O2 over colonies
Oxidase Test (Kovac’s Method ) for bacteria identification
Positive control – Pseudomonas aeruginosa ATCC27853
Negative control – Escherichia coli ATCC 25922
Result
Positive control – Deep violet / purple color
Negative control – No color changes
Test Organism – Deep purple/ violet color
Interpretation
Oxidase positive Organism
In Out
Pseudomonas Escherichia
Aeromonas Salmonella
Plesiomonas Citrobacter
Burkholderia Proteus
Providencia
Morganella
Serratia
Identification
Media required
Hgh and Leifson of media
Peptone water broth
TSI
SIM
Citrate
Urea
Decarboxylase media
MHA
I have passed 3-5 well-isolated colonies from nutrient agar plate to peptone water and incubated at 37 ºC for 2-6 hours for 4-6 m keeping the organisms in log phase for AST.
Also, I have passed a single isolated colony from nutrient agar into 4-6 ml peptone water and incubated it at 37 ºC for 2 -6 hours for getting the organism in the log phase for biochemical tests.
After 4 hours of incubation of peptone water, the broth was compared to 0.5 MacFarland Standard that shows 1.5 x 10^8 CFU/ml( 10^6-10^8 CFU/ml) bacterial cells.
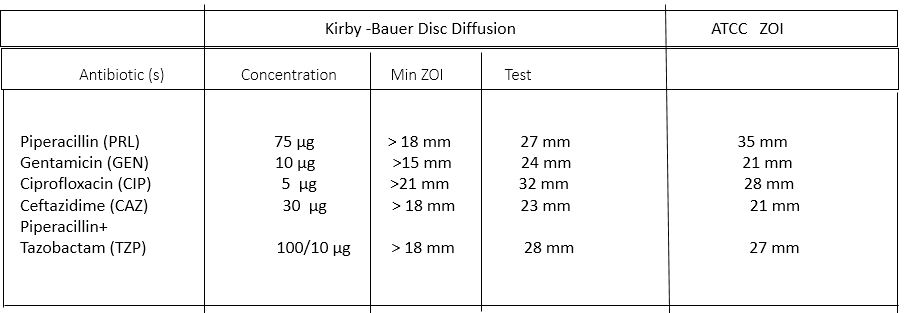
AST is performed on the bacterial isolated by Kirby- Bauer disc diffusion method on MHA.
Purity plate
There is a need for ATCC plates of common organisms like
Staphylococcus aureus ATCC 25923
Enterococcus faecalis ATCC 29912
Pseudomonas aeruginosa ATCC27853
Escherichia coli ATCC 25922
Before inoculation both for biochemical tests and AST
After inoculation both for biochemical and AST
Day 3 for bacterial identification
Oxidative Fermentation test (Hugh and Leifson method )
Pseudomonas aeruginasa ATCC 27853
Open tube (Aerobic ) – Yellow
Overlayer paraffin (Anaerobic ) – Blue-green
Fermentative control
Escherichia coli ATCC 25933
Open tube (Aerobic ) – yellow
Overlayer paraffin ( Anaerobic ) – yellow
oxidizer / Non fermenter reaction
Alkaligens faecalis
Open tube (Aerobic) – green
Overlayed Paraffin ( Anaerobic) – Green
Result
Open / Aerobic tube – yellow
paraffin overlayed – green/non change
organism is oxidative
Triple Sugar Iron Agar
A/A H2S – = Escherichia coli ATCC 25922
K/A H2S + , GAS – =Salmonella enterica serotype Typhi
K/A H2S – GAS + = Salmonella enterica serotype Paratyphi A
k/NC H2S- GAS – = Pseudomonas aeruginosa ATCC 278553
Result: K/NC H2S – Gas –
Sulfide indole motility (SIM) test for bacteria identification
Control
Organisms Indole Motility Sulfide
E. coli + + –
Kleb. pneumoniae – – –
Proteus mirabilis – + +
Test organism – + –
Result = Indole- H2S – Modility +
Citrate utilization test for bacteria identification
Positive control – Klebsiella pneumoniae ATCC 13883
Bluish Color
Negative control – Escherichia coli ATCC 25922
Greenish ( no color change)
Test Organism – Bluish color
Result –Citrate utilization test positive
Decarboxylase Test for bacteria identification
Positive control
lysine – Klebsiella pneumoniae ATCC 13883
ornithine – Enterobacter cloacae ATCC 23355
Arginine – Enterobacter cloacae ATCC 23355
Negative control
Lysine – Enterobacter clocae ATCC 23355
ornithine – Klebsiella pneumoniae ATCC 13883
Arginine – Klebsiella pneumoniae ATCC 13883
Test organism
Lysine = negative
Arginine = positive
Ornithine = negative
In Out
Pseudomonas Burkholderia
Pseudomonas aeruginosa isolated.
Pigment: pyocyanin (yellow-green)
Growth at 42°C
Nitrite reduction test positive
Gelatin liquefaction test positive
Antibiotic Susceptibility Testing
Kirby-Bauer Disc diffusion method
Further Processing
- Epidemiological markers
Pyocin typing
Bacteriophage typing
Serotyping
Restriction fragment length polymorphism (RFLP)
Ribotyping
G+C context = 50 -70 %
Intrinsic resistance
- Ampicillin
- Cotrimoxazole
- Chloramphenicol
- Tetracyclin
Further Readings
- Bailey & Scott’s Diagnostic Microbiology. Editors: Bettey A. Forbes, Daniel F. Sahm & Alice S. Weissfeld, 12th ed 2007, Publisher Elsevier.
- Clinical Microbiology Procedure Handbook Vol. I & II, Chief in editor H.D. Isenberg, Albert Einstein College of Medicine, New York, Publisher ASM (American Society for Microbiology), Washington DC.
- Colour Atlas and Textbook of Diagnostic Microbiology. Editors: Koneman E.W., Allen D.D., Dowell V.R. Jr, and Sommers H.M.
- Cowan & Steel’s Manual for identification of Medical Bacteria. Editors: G.I. Barron & R.K. Felthani, 3rd ed 1993, Publisher Cambridge University Press.
- Jawetz, Melnick and Adelberg’s Medical Microbiology. Editors: Geo. F. Brook, Janet S. Butel & Stephen A. Morse, 21st ed 1998, Publisher Appleton & Lance, Co Stamford Connecticut.
- Mackie and Mc Cartney Practical Medical Microbiology. Editors: J.G. Colle, A.G. Fraser, B.P. Marmion, A. Simmous, 4th ed, Publisher Churchill Living Stone, New York, Melborne, Sans Franscisco 1996.
- Manual of Clinical Microbiology. Editors: P.R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover and R. H. Yolken, 7th ed 2005, Publisher ASM, USA
- Textbook of Diagnostic Microbiology. Editors: Connie R. Mahon, Donald G. Lehman & George Manuselis, 3rd edition2007, Publisher Elsevier.
- Topley & Wilsons Principle of Bacteriology, Virology, and immunology Vol I, II, III, IV & V. Editors: M.T. Parker & L.H. Collier, 8th ed 1990, Publisher Edward Arnold publication, London.
- Medical Microbiology-The Practice of Medical Microbiology Vol-2-12th Edn. –Robert Cruickshank
- District Laboratory Practice in Tropical Countries – Part-2- Monica Cheesebrough- 2nd Edn Update